Academics
| Degree | University/Institution |
|---|---|
| Ph.D. in Plant genomics and molecular biology. | NIPGR, New Delhi |
| M.Sc. in Biotechnology | KIIT University, Bhubaneswar, Odisha |
Work Experience
| Position | University/Organisation | Period |
|---|---|---|
| Scientist C | Institute of Life Sciences, Bhubaneswar | May 2022 - Till Date |
| Research Associate | Institute of Life Sciences, Bhubaneswar | Nov. 2018 - April 2022 |
| Guest Faculty | Ravenshaw University, Cuttack | July 2018 - Nov. 2018 |
| Research Fellow | ICAR-NBAIR, Bangalore | June 2017 - March 2018 |
Awards & Recognition
Research
| Details |
|---|
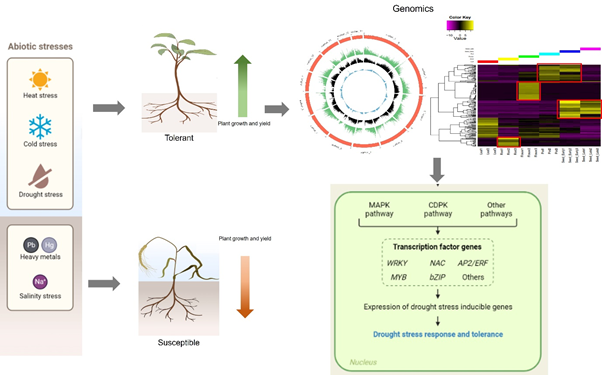
Plants are exposed to many biotic and abiotic stresses every day, which pose a challenge to their growth and development. This ultimately leads to low yield, sup-par seed and fodder quality, and an unstable agro-economy. The rapidly changing climate has compounded this problem and it seems that a food shortage for the expanding world population is imminent. In our lab, we have chosen to study plants’ response to drought and salinity stress, as these account for up to 70% yield loss in major food crops worldwide. We use genomics, transcriptomics, and techniques of molecular biology to understand the effect of these stresses in popular pulses like mung bean (Vigna radiata), moth bean (Vigna aconitifolia), and in native species of minor millets such as little millet (Panicum sumatrense). Millets are marginally grown and consumed in India and have recently been highlighted for their superior nutrient profile compared with rice and wheat. Millets are also naturally climate-resilient and are a good crop for studying plants’ response to drought and salinity since they grow well in nutrient and moisture-poor soils. This also makes them a valuable candidate for future food security. Pulses are an important group of food crops in the world, ranking only after cereals. Many popular pulses like mung bean are susceptible to drought and salinity which affects their yield, leading to heavy losses. However, there are members of the genus Vigna which grow in drought-prone areas and can tolerate higher levels of abiotic stresses like drought and salinity. One such crop is Moth bean (Vigna aconitifolia), which is grown in Rajasthan and Gujarat and is a good resource for studying crop yield under abiotic stress. At our lab, we have used transcriptomics to identify genes that might make little millet and moth bean tolerant to drought and high salinity. Presently, we are integrating the data from whole genome assembly and analysis to identify candidates for functional characterization by knockout and overexpression of selected transcription regulators from these plants in model systems.
|
Publications
| Details |
|---|
Book chapters
|
Group
| Details |
|---|
From left to right: Ms. Udiptanita Rath (JRF), Ms. Samiksha Behera (JRF), Dr. Seema Pradhan, Mr. Biswajit Parida (Lab Technician) |
Grants
Contacts
| Address | Fax | Office | |
|---|---|---|---|
| seema@ils.res.in | Institute of Life Sciences, Nalco Square, Bhubaneswar-751023, India | 0091 674 2300728 |