Genomic Instability and Diseases

Peer reviewed publications:
- Abinash Dutta, Satya Ranjan Sahu, Bhabasha Gyanadeep Utkalaja, Sushree Subhashree Parida, Shraddheya Kumar Patel, Premlata Kumari, and Narottam Acharya (2025) Characterization of dual DNA polymerase knockout strains of Candida albicans with live whole-cell vaccine competence. NPJ Vaccines. 10(1):237. doi: 10.1038/s41541-025-01291-x.
- Satya Ranjan Sahu, Sushree Subhashree Parida, Bhabasha Gyanadeep Utkalaja, Shreenath Nayak, Amrita Dalei, and Narottam Acharya (2025) Enzymatic and structural roles of Candida albicans Rev1 in DNA damage response and disseminated candidiasis. Molecular Microbiology, DOI: 10.1111/mmi.70013.
- Jugal Kishor Sahu, Shweta Thakur, Ipsita Subhadarsini, and Narottam Acharya (2024) p12 isoform-2 is a regulatory subunit of human DNA polymerase delta and is dysregulated in various cancers. FEBS Letters, doi.org/10.1002/1873-3468.15070.
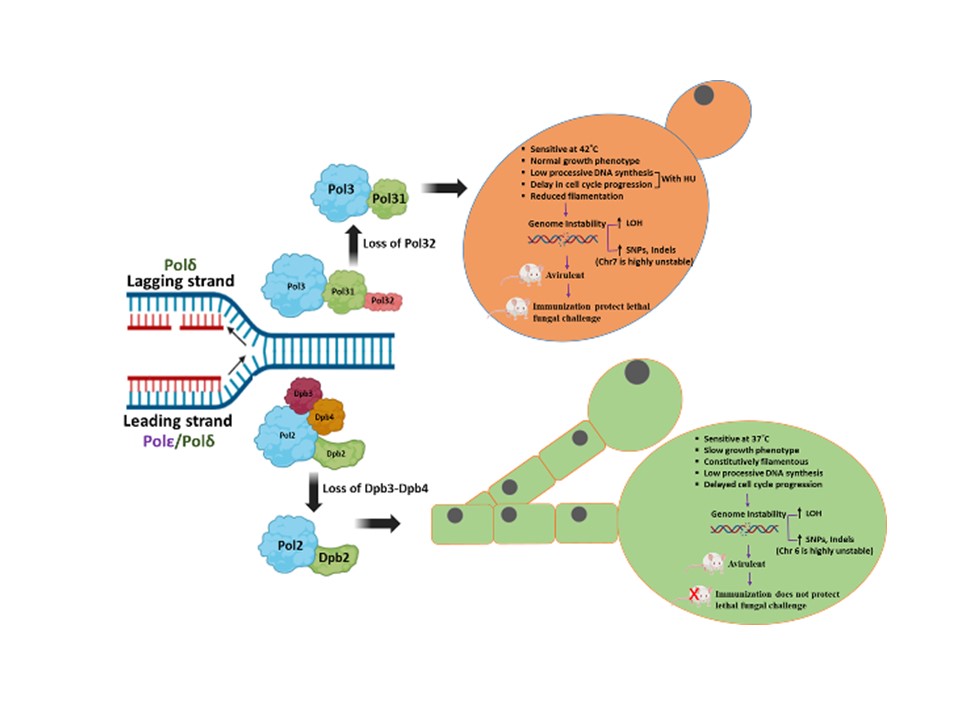
- Bhabasha Gyanadeep Utkalaja, Shraddheya Kumar Patel, Satya Ranjan Sahu, Abinash Dutta, and Narottam Acharya (2024) Critical roles of Dpb3-Dpb4 sub-complex of DNA polymerase epsilon in DNA replication, genome stability, and pathogenesis of Candida albicans. mBio. doi.org/10.1128/mbio.01227-24.
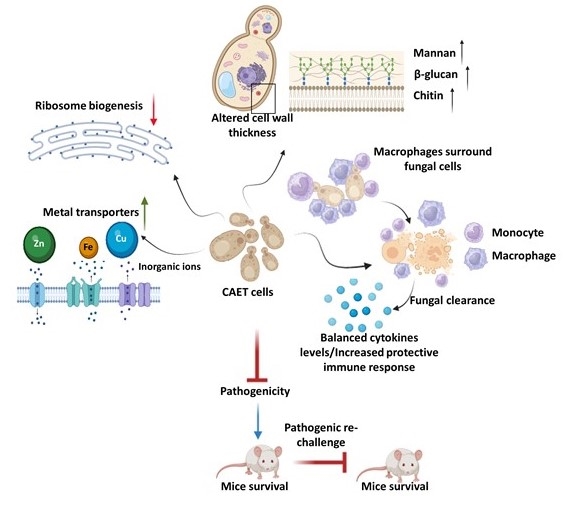
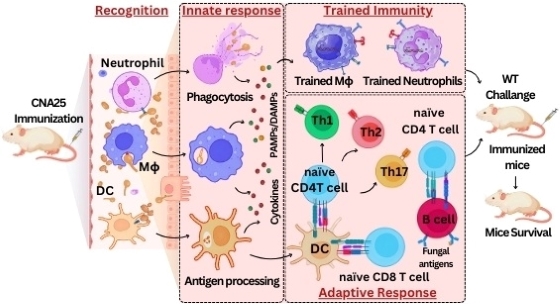
- Satya Ranjan Sahu, Abinash Dutta, Doureradjou Peroumal, Premlata Kumari, Bhabasha Gyanadeep Utkalaja, Shraddheya Kumar Patel, and Narottam Acharya (2024) Immunogenicity and efficacy of CNA25 as a potential whole-cell vaccine against systemic candidiasis. EMBO Molecular Medicine.doi: 10.1038/s44321-024-00080-8.
- Bhabasha Gyanadeep Utkalaja, Satya Ranjan Sahu, Sushree Subhashree Parida, and Narottam Acharya (2024) Hsp90-mediated multi-drug resistance in DNA polymerase defective strains of Candida albicans. Journal of Fungi, 10, 222. https://doi.org/10.3390/jof10030222.
- Swagata Bose, Satya Ranjan Sahu, Abinash Dutta, and Narottam Acharya (2024) A chemically-induced attenuated strain of Candida albicans generates robust protective immune responses and prevents systemic candidiasis development. eLife, 13:RP93760. doi: 10.7554/eLife.93760.
- Satya Ranjan Sahu, Bhabasha Gyanadeep Utkalaja, Shraddheya Kumar Patel, and Narottam Acharya (2023) Spot Assay and Colony Forming Unit (CFU) Analyses-based sensitivity test for Candida albicans and Saccharomyces cerevisiae. Bio-protocol, 13(21):e4872. doi: 10.21769/BioProtoc.4872.
- Satya Ranjan Sahu, Shweta Thakur, Doureradjou Peroumal, Bhabasha Gyanadeep Utkalaja, Abinash Dutta, Premlata Kumari, Ipsita Subhadarsini, and Narottam Acharya (2023) 4-Nitroquinoline 1-oxide induces immune-cells death to onset early immunosuppression during Oral squamous cell carcinoma development. Frontiers in Immunology, 14:1274519. doi: 10.3389/fimmu.2023.1274519.
- Shraddheya Kumar Patel, Satya Ranjan Sahu, and Narottam Acharya (2023) Cell cycle Analysis of Candida albicans by Flow Cytometry. Bio-protocol, 13(20): e4748. DOI: 10.21769/BioProtoc.4748.
- Premlata Kumari, Satya Ranjan Sahu, Abinash Dutta, Bhabasha Gyanadeep Utkalaja, and Narottam Acharya (2023) RAD51-WSS1 dependent genetic pathways are essential for DNA-Protein crosslink repair and pathogenesis in Candida albicans. Journal of Biological Chemistry, doi.org/10.1016/j.jbc.2023.104728. (On the cover page)
- Swagata Bose, Durg Vijai Singh, Tapan Kumar Adhya, and Narottam Acharya (2023) Escherichia coli, but Not Staphylococcus aureus, Functions as a Chelating Agent That Exhibits Antifungal Activity against the Pathogenic Yeast Candida albicans. Journal of Fungi, 9, 286. https://doi.org/10.3390/jof9030286
- Shraddheya Kumar Patel, Satya Ranjan Sahu, Bhabasha Gyanadeep Utkalaja, Swagata Bose, and Narottam Acharya (2023) Pol32, an accessory subunit of DNA polymerase delta, plays an essential role in genome stability and pathogenesis of Candida albicans. Gut microbes, DOI: 10.1080/19490976.2022.2163840.
- Narottam Acharya, Louise Prakash, and Satya Prakash (2023) Yeast 9-1-1 complex acts as a sliding clamp for DNA synthesis by DNA polymerase ε. Journal of Biological Chemistry, Jan;299(1):102727. doi: 10.1016/j.jbc.2022.102727.
- Doureradjou Peroumal, Satya Ranjan Sahu, Premlata Kumari, Bhabasha G. Utkalaja, and Narottam Acharya (2022) Commensal Fungus Candida albicans Maintains a Long-Term Mutualistic Relationship with the Host To Modulate Gut Microbiota and Metabolism. Microbiology Spectrum, DOI: 10.1128/spectrum.02462-22.
- Satya Ranjan Sahu, Swagata Bose, Manish Singh, Premlata Kumari, Abinash Dutta, Bhabasha G. Utkalaja, Shraddheya Kumar Patel, and Narottam Acharya (2022) Vaccines against Candidiasis: Status, Challenges and Emerging opportunity. Front. Cell. Infect. Microbiol., DOI: 10.3389/fcimb.2022.1002406.
- Kodavati Manohar, Prashant Khandagale, Shraddheya Kumar Patel, Jugal Kishor Sahu, and Narottam Acharya (2021) Ubiquitin-binding domain of DNA polymerase eta lacking the PCNA-interacting-protein motif directly binds to PCNA and regulates translesion DNA synthesis. Journal of Biological Chemistry, Dec 18;298(2):101506. doi: 10.1016/j.jbc.2021.101506.
- Premlata Kumari, Rajivgandhi Sundaram, Kodavati Manohar, Dileep Vasudevan and Narottam Acharya (2021) Interdomain connecting loop and J loop structures determine cross-species compatibility of PCNA. Journal of Biological Chemistry, Jul;297(1):100911. doi: 10.1016/j.jbc.2021.100911.
- Rajivgandhi Sundaram, Kodavati Manohar, Shraddheya Kumar Patel, Narottam Acharya and Dileep Vasudevan (2021) Structural analyses of PCNA from the fungal pathogen Candida albicans. FEBS Letters, May;595(9):1328-1349. doi: 10.1002/1873-3468.
- Narottam Acharya, Shraddheya Kumar Patel, Satya Ranjan Sahu, and Premlata Kumari (2020) “PIPs” in DNA polymerase – PCNA interaction affairs. Biochem. Soci. Trans. Nov 16:BST20200678. doi: 10.1042/BST20200678.
- Swagata Bose, Shifu Aggarwal, DurgVijai Singh and Narottam Acharya (2020) Extracellular vesicles: An emerging platform in gram-positive bacteria. Micro. cell Vol. 7, No. 12, pp. 312 – 322; doi: 10.15698/mic2020.12.737.
- Prashant Khandagale, Shweta Thakur and Narottam Acharya (2020) Identification of PCNA interacting protein motifs in human DNA polymerase delta, Bioscience Reports, 40 (4). (doi.org/10.1042/BSR20200602)
- Narottam Acharya, Prashant Khandagale, Shweta Thakur, Jugal Kishor Sahu and Bhabasha Gyanadeep Utakalaja (2020) Quaternary structural diversity in Eukaryotic DNA Polymerases: Monomeric to Multimeric form. Current Genetics, Aug; 66 (4):635-655. doi: 10.1007/s00294-020-01071-1.
- Doureradjou Peroumal, Kodavati Manohar, Shraddheya Kumar Patel, Premlata Kumari, Satya Ranjan Sahu and Narottam Acharya (2019) Virulence and pathogenicity of a Candida albicans mutant with reduced filamentation. Cellular Microbiology, 21 (12) (doi.org/10.1111/cmi.13103).
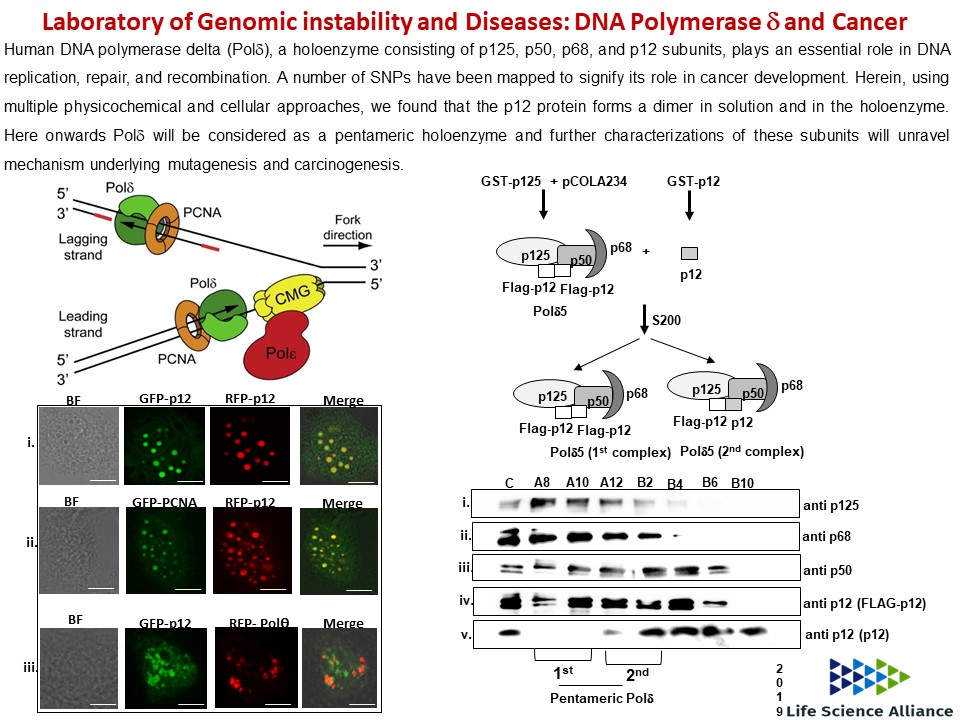
- Prashant Khandagale, Doureradjou Peroumal, Kodavati Manohar and Narottam Acharya (2019) Human DNA polymerase delta is a pentameric holoenzyme with a dimeric p12 subunit. Life Science Alliance, Vol. 2, No., 2. (doi.org/10.26508/lsa.201900323)
- Narottam Acharya, Kodavati Manohar, D. Peroumal, Prashant Khandagale, Shraddheya Kumar Patel, Satya Ranjan Sahu, and Premlata Kumari (2019) Multifaceted activities of DNA Polymerase eta: Beyond translesion DNA synthesis. Curr. Genetics, 65(3), 649-656. (doi.org/10.1007/s00294-018-0918-5).
- Jingde, N., Ray, R., Sinha, S., Rana, K., Singh, S. K., Khandagale, P., Narottam Acharya and V. Rai (2018) Cysteine mediated disulfide bond formation in RAGE V domain facilitates its functionally relevant dimerization. Biochimie, 154, 55-61.
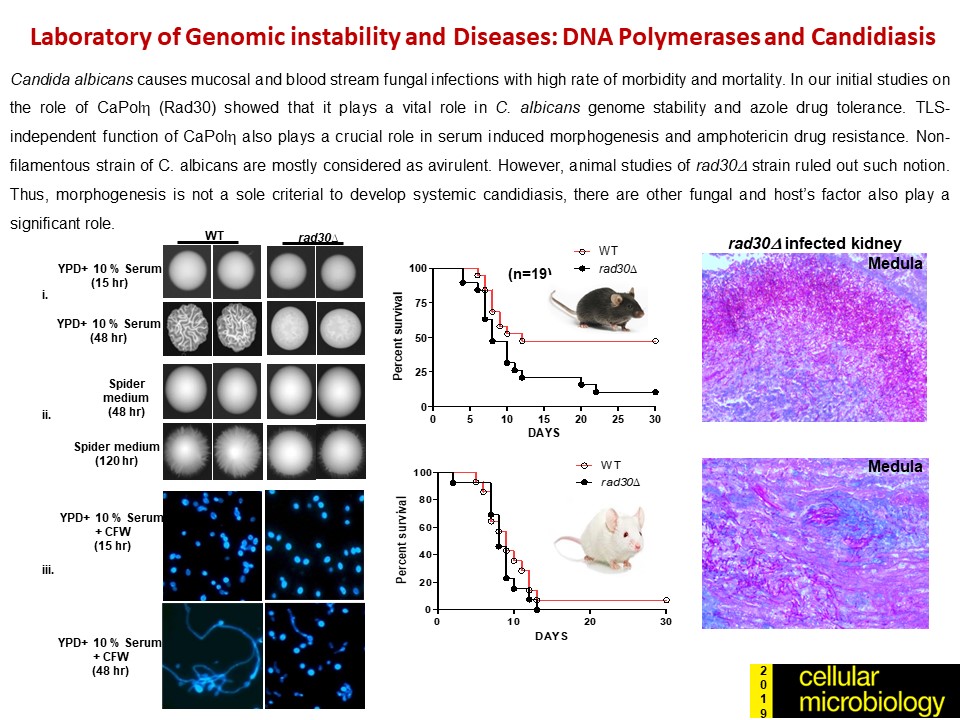
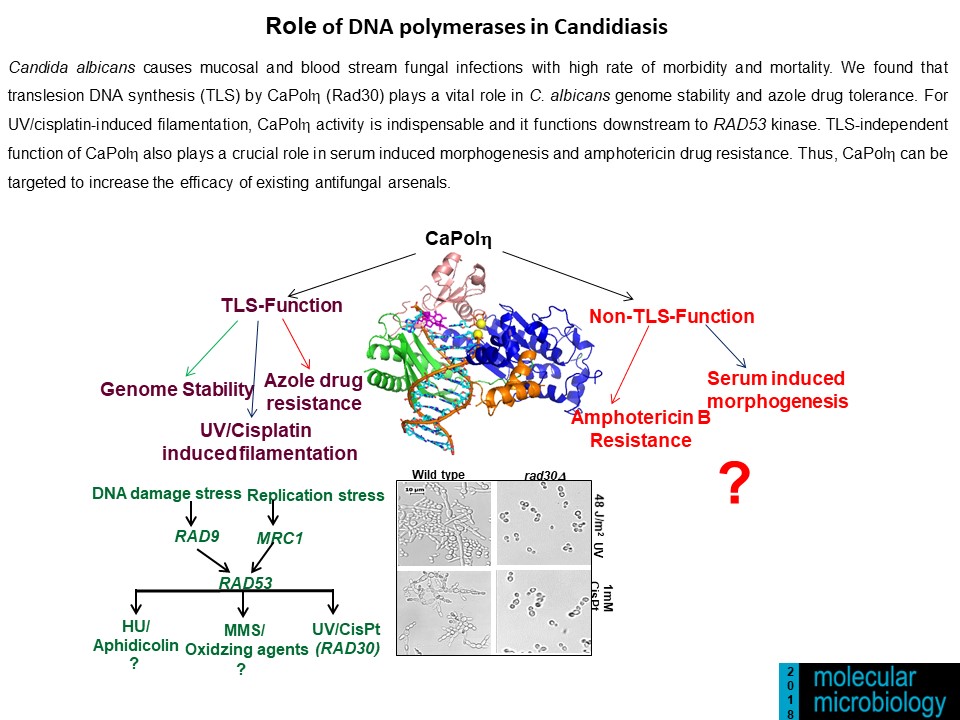
- Kodavati Manohar, Doureradjou Peroumal and Narottam Acharya (2018) TLS dependent and independent functions of DNA polymerase eta (Polh/ Rad30) from Pathogenic Yeast Candida albicans. Molecular Microbiology, 110 (5), 707-727
- Suresh Satpati, Kodavati Manohar, Narottam Acharya*, Anshuman Dixit. (2017) Comparative molecular dynamics studies of heterozygous open reading frames of DNA polymerase eta (η) in pathogenic yeast Candida albicans. Sci Rep., 7:41087. doi: 10.1038/srep41087. (Joined corresponding author)
- Kodavati Manohar and Narottam Acharya (2015) Characterization of Proliferating Cell Nuclear Antigen (PCNA) from Pathogenic Yeast Candida albicans and its functional analyses in S. cerevisiae. BMC-Microbiology, 15:257.
- Yoon J.H+., Acharya N+., Jeseong P., Debashree Basu, Prakash S. and Prakash L. (2014) Identification of two functional PCNA binding domains in human DNA polymerase κ (+ joint first author). Genes to Cells., 19(7):594-601.
- Acharya N., Kalssen R., Johnson R.E., Prakash L. and Prakash S. (2011) PCNA binding domains in all three subunits of yeast DNA polymerase δ modulate its function in DNA replication. Proc Natl Acad Sci USA., Nov.1; 108(44):17927-17932.
- Acharya N+., Yoon J.H+., Hutwitz J., Prakash L. and Prakash S. (2010) DNA polymerase eta lacking the ubiquitin-binding domain promotes replicative lesion bypass in human cells (+ joint first author). Proc Natl Acad Sci USA., Jun 8; 107(23):10401-10405.
- Acharya N., Johnson R.E., Pagès V., Prakash L. and Prakash S. (2009) Yeast Rev1 protein promotes complex formation of DNA polymerase zeta with Pol32 subunit of DNA polymerase delta. Proc Natl Acad Sci USA., Jun 16; 106(24):9631-9636.
- Acharya N., Yoon J.H., Gali H., Unk I., Haracska L., Johnson R.E., Hutwitz J., Prakash L. and Prakash S. (2008) Roles of PCNA-binding and ubiquitin-binding domains in human DNA polymerase eta in translesion DNA synthesis. Proc Natl Acad Sci USA., Nov 18; 105 (46):17724-17729.
- Pagès V., Bresson A., Acharya N., Prakash L. Fuchs R.P. and Prakash S. (2008) Requirement of Rad5 for DNA polymerase zeta-dependent translesion synthesis in Saccharomyces cerevisiae. Genetics, 2008 Sep; 180(1):73-82.
- Acharya N., Haracska L., Prakash S. and Prakash L. (2007) Complex formation of yeast Rev1 with DNA polymerase {eta}. Mol Cell Biol., Dec; 27 (23):8401-8408.
- Acharya N., Brahma A., Haracska L., Prakash S. and Prakash L. (2007) Mutations in the Ubiquitin Binding UBZ Motif of DNA Polymerase {eta} Do Not Impair Its Function in Translesion Synthesis during Replication. Mol Cell Biol., Oct; 27(20):7266-7272.
- Acharya N., Johnson R.E., Prakash S. and Prakash L. (2006) Complex formation with Rev1 enhances the proficiency of Saccharomyces cerevisiae DNA polymerase zeta for mismatch extension and for extension opposite from DNA lesions. Mol. Cell Biol., Dec. 26 (24), 9555-9563.
- Acharya N., Haracska L., Johnson R.E., Unk I., Prakash S. and Prakash L. (2005) Complex formation of yeast Rev1 and Rev7 proteins: a novel role for the polymerase-associated domain. Mol. Cell Biol., Nov. 25 (21), 9734-9740.
- Haracska L., Acharya N., Unk I., Johnson R.E., Hurwitz J., Prakash L. and Prakash S. (2005) A single domain in human DNA polymerase iota mediates interaction with PCNA: implications for translesion DNA synthesis. Mol. Cell Biol., Feb. 25 (3), 1183-1190
- Acharya N, Talawar R. K., Purnapatre K, Varshney U. (2004) Use of sequence microdivergence in mycobacterial ortholog to analyze contributions of the water-activating loop histidine of Escherichia coli uracil-DNA glycosylase in reactant binding and catalysis. Biochem. Biophys. Res. Commun., 320(3), 893-899.
- Acharya, N., Talawar R. K., Saikrishnan, K., Vijayan, M and Varshney, U. (2003) Substitutions at tyrosine 66 of Escherichia coli uracil DNA glycosylase lead to characterization of an efficient enzyme that is recalcitrant to product inhibition. Nucleic Acids Res., 31(24), 7216-7226.
- Acharya, N., Kumar P. and Varshney, U. (2003) Complexes of Uracil-DNA glycosylase inhibitor protein, Ugi, with Mycobacterium smegmatis and Mycobacterium tuberculosis uracil-DNA glycosylases. Microbiology 149, 1647-1658.
- Saikrishnan, K., Jeyakanthan, Venkatesh, J., Acharya, N., Purnapatre, K., Sekar, K., Varshney, U. and Vijayan, M. (2003) Structure of Mycobacterium tuberculosis single stranded DNA binding protein. Variability in quaternary structure and its implications. J. Mol. Biol., 331, 385-393.
- Acharya, N., Roy, S., and Varshney, U. (2002) Mutational analysis of the uracil DNA glycosylase inhibitor protein and its interaction with Escherichia coli uracil DNA glycosylase. J. Mol. Biol., 321 (4), 579-590.
- Acharya, N., and Varshney, U. (2002) Biochemical properties of single stranded DNA binding protein from Mycobacterium smegmatis, a fast growing mycobacterium and its physical and functional interaction with uracil DNA glycosylases. J. Mol. Biol., 318 (5), 1251-1264.
- Handa, P.+, Acharya, N.+, and Varshney, U. (2002) Effects of mutations at tyrosine 66 and asparagine 123 in the active site pocket of Escherichia coli uracil DNA glycosylase on uracil excision from synthetic DNA oligomers: evidence for the occurrence of long-range interactions between the enzyme and substrate. (+ joint first author) Nucleic Acids Res, 30(14), 3086-3095.
- Handa, P., Acharya, N., Talwar R. K., Roy, S., and Varshney, U. (2002) Contrasting effects of mutating active site residues, aspartic acid 64 and histidine 187 of Escherichia coli uracil-DNA glycosylase on uracil excision and interaction with an inhibitor protein. Indian J. Biochem. Biophys., 29, 312-317.
- Saikrishnan, K., Jeyakanthan, Venkatesh, J., Acharya, N., Purnapatre, K., Sekar, K., Varshney, U. and Vijayan, M. (2002) Crystallization and preliminary X-ray studies on the single stranded DNA binding protein from Mycobacterium tuberculosis. Acta Crystallogr D Biol Crystallogr, 58, 327-329.
- Handa, P., Acharya, N. and Varshney, U. (2001) Chimeras between single stranded DNA binding proteins from Escherichia coli and Mycobacterium tuberculosis reveal that their C-terminal domains interact with uracil-DNA glycosylases. J. Biol. Chem. 276, 16992-16997.
- Handa, P., Acharya, N., Thanedar, S., Purnapatre, K. and Varshney, U. (2000) Distinct properties of single stranded DNA binding protein from Mycobacterium tuberculosis and its functional characterization in Escherichia coli. Nucleic Acids Res. 28, 3823-3829.
- Sadhale, P., Sharma, N., Beena, P, Katoch, A., Acharya, N., and Singh, S. K. (1998) Modulation of polymerase II composition: a possible mode of transcriptional regulation of stress response in eukaryotes. J. Biosci. 23, 331-335.
Pre-prints
- Swagata Bose, Satya Ranjan Sahu, Abinash Dutta, Narottam Acharya (2023) A chemically-induced attenuated strain of Candida albicans generates robust protective immune responses and prevents systemic candidiasis development. bioRxiv 2023.11.03.565495; doi: https://doi.org/10.1101/2023.11.03.565495
- Doureradjou Peroumal, Satya Ranjan Sahu, Premlata Kumari, Bhabasha Utkalaja, Narottam Acharya (2022) Commensal fungi Candida albicans modulates dietary high-fat induced alterations in metabolism, immunity, and gut microbiota. bioRxiv 2022.03.23.485455; doi: https://doi.org/10.1101/2022.03.23.485455.
- Prashant Khandagale, Shweta Thakur, Narottam Acharya (2020) Identification of PCNA interacting protein motifs in human DNA polymerase delta. bioRxiv 2020.02.29.971473; doi: https://doi.org/10.1101/2020.02.29.971473.
- Prashant Khandagale, Doureradjou Peroumal, Kodavati Manohar, Narottam Acharya (2019) Human DNA polymerase delta is a pentameric holoenzyme with dimeric p12 subunit: Implications in enzyme architecture and PCNA interaction. bioRxiv 525485; doi: https://doi.org/10.1101/525485.
Book chapter
- Narottam Acharya*, Kodavati Manohar, Shreenath Nayak, Avishek Chatterjee and Amrita Dalei (2106) DNA Polymerase: A putative drug target against Candidiasis. Frontiers in Life Sciences, Excel Indian Publishers, New Delhi, India. Pp. 12-23
|


























 EMBO MOLECULAR MEDICENE- 2024
EMBO MOLECULAR MEDICENE- 2024