Academics
| Degree | University/Institution |
|---|---|
| Ph.D. Molecular Biology, Microbiology and Biochemistry | Molecular Biology, Microbiology and Biochemistry |
| M.Sc. Biotechnology | M.S University, Baroda, Gujarat, India |
Work Experience
| Position | University/Organisation | Period |
|---|---|---|
| Scientist E | Institute of Life Sciences, Bhubaneswar, Orissa | July 2021 - Till date |
| Scientist D | Institute of Life Sciences, Bhubaneswar, Orissa | March 2016 - June 2020 |
| Post-Doctoral fellow | Dept. of Bioscience and Nutrition, Karolinska Institutet, Stockholm, Sweden | September 2010 - March 2016 |
| Junior Research Fellow | Dept. of Microbiology and Cell Biology, Indian Institute of Science, Bangalore, India | June 2003 - July 2004 |
| Researcher | Avesthagen Pvt. Ltd, Bangalore, India | August 2000 - May 2003 |
Awards & Recognition
| Details |
|---|
|
Research
| Details |
|---|
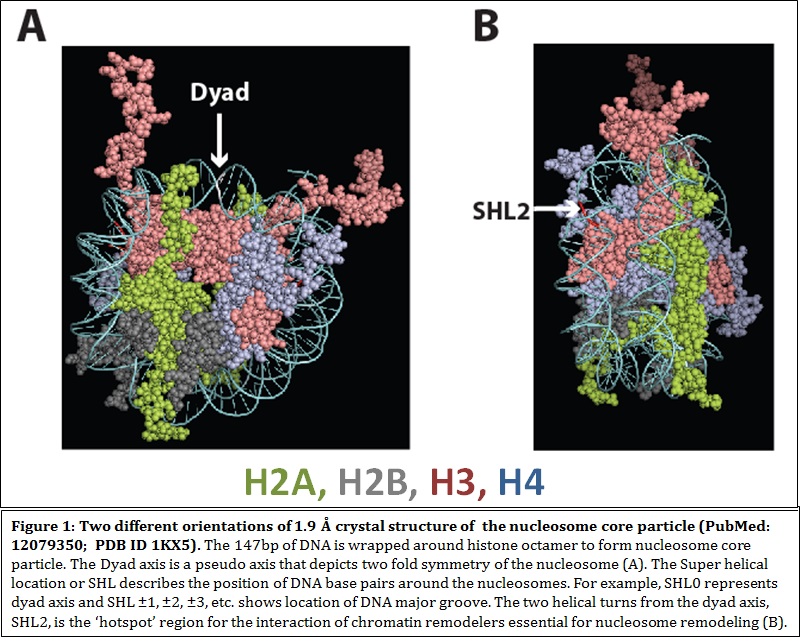
Characterization of chromatin remodelers and epigenetic factors in blood cell development How a single cell can divide and differentiate into an entire organism is of immense intrigue to me. In an organism, cells, tissues and organs work in concert for normal development, perturbation of which causes developmental disorders and disease. My laboratory is interested to find out the contribution of chromatin remodeling complexes and their subunits towards development and diseases of the human hematopoietic system. Chromatin and chromatin remodelers The highly compacted eukaryotic genome is composed of nucleosomes as its basic unit. Nucleosomes are composed of 147 bp of DNA wrapped around an octameric core of histone proteins. To make DNA accessible for factors regulating biological processes, cells possess enzymes called ATP-dependent chromatin remodeling complexes (CRCs). These enzymes utilize the energy of ATP hydrolysis to remodel chromatin. CRCs belong to the SNF2 family of ATPases and have been classified into subfamilies based on the presence of different domains, other than the ATPase domain present in the catalytic subunit. The four families of CRCs are SWI/SNF (SWItch/Sucrose Non-fermenting), ISWI (Imitation Switch), CHD (Chromodomain helicase DNA binding proteins)and INO80/SWR1. CRCs, which are usually multisubunit enzyme complexes, possess several domains in its ATPase and auxiliary subunits, which interpret epigenetic codes residing on histone tails, thereby determining the fate of chromatin and gene expression.
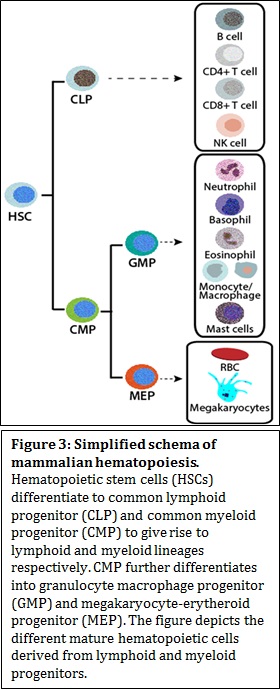
Decades of research from various laboratories have shown that CRCs have distinct and redundant ways to change chromatin architecture, namely, by nucleosome assembly, sliding, spacing and eviction, dimer displacement and change of nucleosome composition with histone variants. To add another level of complexity, CRCs have tissue-specific or developmental stage specific isoforms, which determines the development or lineage commitment of the tissue/cell type in question. In other words, a combinatorial association of auxiliary subunits can define the lineage choice. Impaired function of CRCs has been implicated in developmental disorders and cancer. Mammalian hematopoiesis In hematopoiesis, various types of blood cells are produced from hematopoietic stem cells (HSCs) in the bone marrow. Hematopoietic differentiation can be broadly categorized into two major lineages; lymphoid and myeloid lineages. Lymphoid lineage gives rise to mature B cells, T cells, and natural killer cells and the myeloid lineage give rise to granulocytes, (consisting of neutrophils, basophils, eosinophils) monocytes, macrophages, megakaryocytes, platelets, and erythrocytes. Differentiation block in myeloid progenitors results in an abnormal clonal expansion of myeloid blast cells in peripheral blood and in bone marrow. The blast cells fail to undergo normal maturation process to produce different kinds of normal myeloid cells and result in a population of myeloid blasts. Myeloid blasts or leukemic blasts over populate normal myeloid cells thereby compromising an individual’s normal blood homeostasis. Such abnormalities in myeloid differentiation result in myeloid malignancies called Acute Myeloid Leukemia (AML). AML is a heterogeneous disease that consists of several subtypes based on cytogenetic and genetic alterations, which is also the basis for classification and prognosis. AML is characterized by disturbed transcriptional regulation that leads to a differentiation block and increased proliferation. To determine the abnormalities in AML, it is, therefore, important to understand normal myelopoiesis.
The research interest of the lab is identification and characterization of different chromatin remodelers and their subunits towards development and diseases of the human hematopoietic system. We are specifically interested in understanding myeloid development/differentiation program and how impairment in gene expression results in myeloid disorders. We use molecular biology, biochemistry, and next-generation sequencing approaches to understand how chromatin remodelers regulate chromatin architecture which in turn regulate gene expression for myeloid lineage choice and differentiation. |
Publications
| Details |
|---|
2024
2023
2022
2021
2020
2019
2015
2014
2013
2012
2009
Book Chapters
Reviews
|
Group
Grants
| Details |
|---|
Generous intramural start-up support from ILS Ramalingaswami Fellowship – 2016 SERB EMR – 2019-2022 ICMR – 2022-2025 |
Contacts
| Address | Fax | Office | |
|---|---|---|---|
| punit@ils.res.in | Institute of Life Sciences, Nalco Square, Chandrasekharpur, Bhubaneswar, Odisha-751023 India | +91 674 2300728 | +91 674 2304319 |
Highlights
| Details | ||
|---|---|---|
Cover Pages
|
Positions
| Details |
|---|
My lab is actively looking for prospective enthusiastic Post Doctoral fellows, who are interested in the field of chromatin remodeling, epigenetics and hematopoiesis. Scholars who have finished/finishing their Ph.D. are encouraged to apply with their detailed CV and one to two pages write-up describing their research interests. Candidates with experience in molecular biology, cell biology and bioinformatics will be preferred. The applicants should have postdoctoral fellowship or willing to apply for the fellowship. I will be happy to guide prospective candidates through the research proposal and application process for obtaining Postdoc fellowships. Vacancy for a SRF position under SERB EMR (2019-2022) project for three years. Looking for a person with strong bioinformatics background and knowledge of biological sciences. Interested candidates should contact me at punitprasad.ils@gmail.com; punit@ils.res.in |